
Breaking News
 Iran (So Far Away) - Official Music Video
Iran (So Far Away) - Official Music Video
 COMEX Silver: 21 Days Until 429 Million Ounces of Demand Meets 103 Million Supply. (March Crisis)
COMEX Silver: 21 Days Until 429 Million Ounces of Demand Meets 103 Million Supply. (March Crisis)
 Marjorie Taylor Greene: MAGA Was "All a Lie," "Isn't Really About America or the
Marjorie Taylor Greene: MAGA Was "All a Lie," "Isn't Really About America or the
 Why America's Two-Party System Will Never Threaten the True Political Elites
Why America's Two-Party System Will Never Threaten the True Political Elites
Top Tech News
 How underwater 3D printing could soon transform maritime construction
How underwater 3D printing could soon transform maritime construction
 Smart soldering iron packs a camera to show you what you're doing
Smart soldering iron packs a camera to show you what you're doing
 Look, no hands: Flying umbrella follows user through the rain
Look, no hands: Flying umbrella follows user through the rain
 Critical Linux Warning: 800,000 Devices Are EXPOSED
Critical Linux Warning: 800,000 Devices Are EXPOSED
 'Brave New World': IVF Company's Eugenics Tool Lets Couples Pick 'Best' Baby, Di
'Brave New World': IVF Company's Eugenics Tool Lets Couples Pick 'Best' Baby, Di
 The smartphone just fired a warning shot at the camera industry.
The smartphone just fired a warning shot at the camera industry.
 A revolutionary breakthrough in dental science is changing how we fight tooth decay
A revolutionary breakthrough in dental science is changing how we fight tooth decay
 Docan Energy "Panda": 32kWh for $2,530!
Docan Energy "Panda": 32kWh for $2,530!
 Rugged phone with multi-day battery life doubles as a 1080p projector
Rugged phone with multi-day battery life doubles as a 1080p projector
 4 Sisters Invent Electric Tractor with Mom and Dad and it's Selling in 5 Countries
4 Sisters Invent Electric Tractor with Mom and Dad and it's Selling in 5 Countries
Nearly 100% of bacterial infections can now be identified in under 3 hours
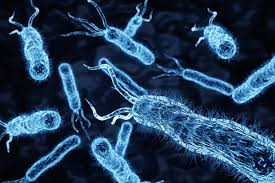
Scientists at the Ulsan National Institute of Science & Technology (UNIST) in South Korea have unveiled their novel diagnostic technique known as fluorescence in situ hybridization (FISH), using artificial polymers – peptide nucleic acid (PNA) – that act as probes to bind to different genetic sequences within bacteria. When the two probe molecules bind to the target, fluorescent signals are emitted, which essentially reveal the fingerprint of different pathogens.
"The fluorescence in situ hybridization (FISH) technique allows the rapid detection and identification of microbes based on their variation in genomic sequence without time-consuming culturing or sequencing," the scientists noted. "However, the recent explosion of microbial genomic data has made it challenging to design an appropriate set of probes for microbial mixtures. We developed a novel set of peptide nucleic acid (PNA)-based FISH probes with optimal target specificity by analyzing the variations in 16S ribosomal RNA sequence across all bacterial species."



